https://ies.ed.gov/ncee/wwc/Docs/referenceresources/WWC_Procedures_Handbook_V4_1_Draft.pdf
See Page 16.
two_values<-view1b[3:4]
num<-view1b[2]
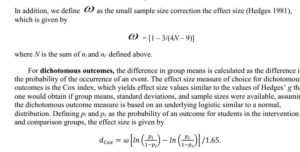
omega<-(1-3/(4*num-9))
C_LOGIT<-two_values[1]
C_EXP<-exp(C_LOGIT)/(1+exp(C_LOGIT))
C_ODDS<-C_EXP/(1-C_EXP)
C_STEP1<-log(C_ODDS)
T_LOGIT<-two_values[1]+two_values[2]
T_EXP<-exp(T_LOGIT)/(1+exp(T_LOGIT))
T_ODDS<-T_EXP/(1-T_EXP)
T_STEP1<-log(T_ODDS)
STEP2<-T_STEP1-C_STEP1
wwc_effect_size<-STEP2/1.65
wwc_effect_size_omega<-(omega*STEP2)/1.65
odds_ratio<-T_ODDS/C_ODDS